Getting Started – DIA Preprocesing
DO-MS is an application to visualize mass-spec data both in an interactive application and static reports generated via. the command-line. In this document we’ll walk you through analyzing an example dataset in the interactive application.
Table of Contents
Example Data
We have provided an example data set online, which contains parts of the MS2 number optimization in the paper. You can download a .zip bundle of it here: https://drive.google.com/file/d/1bjFzKqTFLk7ECUJOTy8LNxsCOD0Xf96Q/view?usp=share_link. The contents of the archive are the main dia-nn report and the corresponding raw files.
Your folder should look like this:
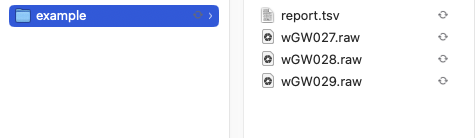
Installation
Please make sure that you installed DO-MS as descibed in the installation section. For using the preprocessing pipeline it is necessary to install the ThermoRawFileParser and the Dinosaur feature detection.
Processing Raw Data
Open a terminal and enter the base folder of your DO-MS installation. Make sure that your DO-MS environment is set up and activate it.
conda activate doms
For processing, the piplline module located at pipeline/processing.py will be called with the following parameters.
python pipeline/processing.py /location/to/example/report_filtered.tsv
the following additional options will be included:
# Activate Mono if using Mac or Linux. Mono is required to run the Thermo Raw File Parser on Linux and OSX.
-m
# location of the ThermoRawFileParser executeable
--raw-parser-location /location/to/ThermoRawFileParser1.4.2/ThermoRawFileParser.exe
# location of the Dinosaur .jar file
--dinosaur-location /Users/georgwallmann/Library/CloudStorage/OneDrive-Personal/Studium/Northeastern/DO-MS-DIA/Dinosaur-1.2.0.free.jar
# location of the example raw data
-r /location/to/example
The full command needs to be a single line and will look like:
python pipeline/processing.py /location/to/example/report_filtered.tsv -m --raw-parser-location /location/to/ThermoRawFileParser1.4.2/ThermoRawFileParser.exe --dinosaur-location /Users/georgwallmann/Library/CloudStorage/OneDrive-Personal/Studium/Northeastern/DO-MS-DIA/Dinosaur-1.2.0.free.jar -r /location/to/example
After processing, the additional files should be part of your folder:
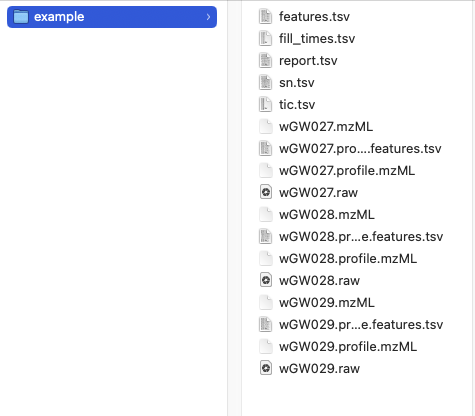
Temporary .mzML files can be deleted.
Command Line Interface
The documentation for the various command line options can be found by typing python pipeline/processing.py -h
usage: processing.py [-h] --raw-parser-location RAW_PARSER_LOCATION
[--dinosaur-location DINOSAUR_LOCATION] [-m] [-d] [-v]
[-t TEMPORARY_FOLDER] [-r RAW_FILE_LOCATION]
[--no-feature-detection] [--no-fill-times] [--no-tic]
[--no-sn] [--no-mzml-generation]
[--mz-bin-size MZ_BIN_SIZE] [--rt-bin-size RT_BIN_SIZE]
[--resolution RESOLUTION] [-p PROCESSES] [--isotopes-sn]
report
Command line tool for feature detection in shotgun MS experiments. Can be used
together with DIA-NN to provide additional information on the peptide like
features identified in the MS1 spectra.
positional arguments:
report Location of the report.tsv output from DIA-NN which
should be used for analysis.
options:
-h, --help show this help message and exit
--raw-parser-location RAW_PARSER_LOCATION
Path pointing to the ThermoRawFileParser executeable.
--dinosaur-location DINOSAUR_LOCATION
Path pointing to the dinosaur jar executeable.
-m, --mono Use mono for ThermoRawFileParser under Linux and OSX.
-d, --delete Delete generated mzML and copied raw files after
successfull feature generation.
-v, --verbose Show verbose output.
-t TEMPORARY_FOLDER, --temporary-folder TEMPORARY_FOLDER
Input Raw files will be temporarilly copied to this
folder. Required for use with Google drive.
-r RAW_FILE_LOCATION, --raw-file-location RAW_FILE_LOCATION
By default, raw files are loaded based on the
File.Name column in the report.tsv. With this option,
a different folder can be specified.
--no-feature-detection
All steps are performed as usual but Dinosaur feature
detection is skipped. No features.tsv file will be
generated.
--no-fill-times All steps are performed as usual but fill times are
not extracted. No fill_times.tsv file will be
generated.
--no-tic All steps are performed as usual but binned TIC is not
extracted. No tic.tsv file will be generated.
--no-sn Signal to Noise ratio is not estimated for precursors
--no-mzml-generation Raw files are not converted to .mzML. Nevertheless,
mzML files are expected in their theoretical output
location and loaded. Should be only be carefully used
for repeated calulcations or debugging
--mz-bin-size MZ_BIN_SIZE
Bin size over the mz dimension for TIC binning.
--rt-bin-size RT_BIN_SIZE
Bin size over the RT dimension for TIC binning in
minutes. If a bin size of 0 is provided, binning will
not be applied and TIC is given per scan.
--resolution RESOLUTION
Set the resolution used for estimating counts from S/N
data
-p PROCESSES, --processes PROCESSES
Number of Processes
--isotopes-sn Use all isototopes from the same scan as the highest
intensity datapoint for estimating the SN and copy
number.